5 Pull it Together
We write papers. Those papers cite books and articles. They often incorporate tables and figures created in R. What we want to do is quickly turn a Markdown file containing things like that into a properly formatted scholarly paper, without giving up any of the necessary scholarly apparatus (on the output side) or the convenience and convertibilty of Markdown (on the input side). We want to easily get good-looking output from the same source in HTML, PDF, and DOCX formats. And we want to do that with an absolute minimum of—ideally, no—post-processing of the output beyond the basic conversion step. This is within our reach.
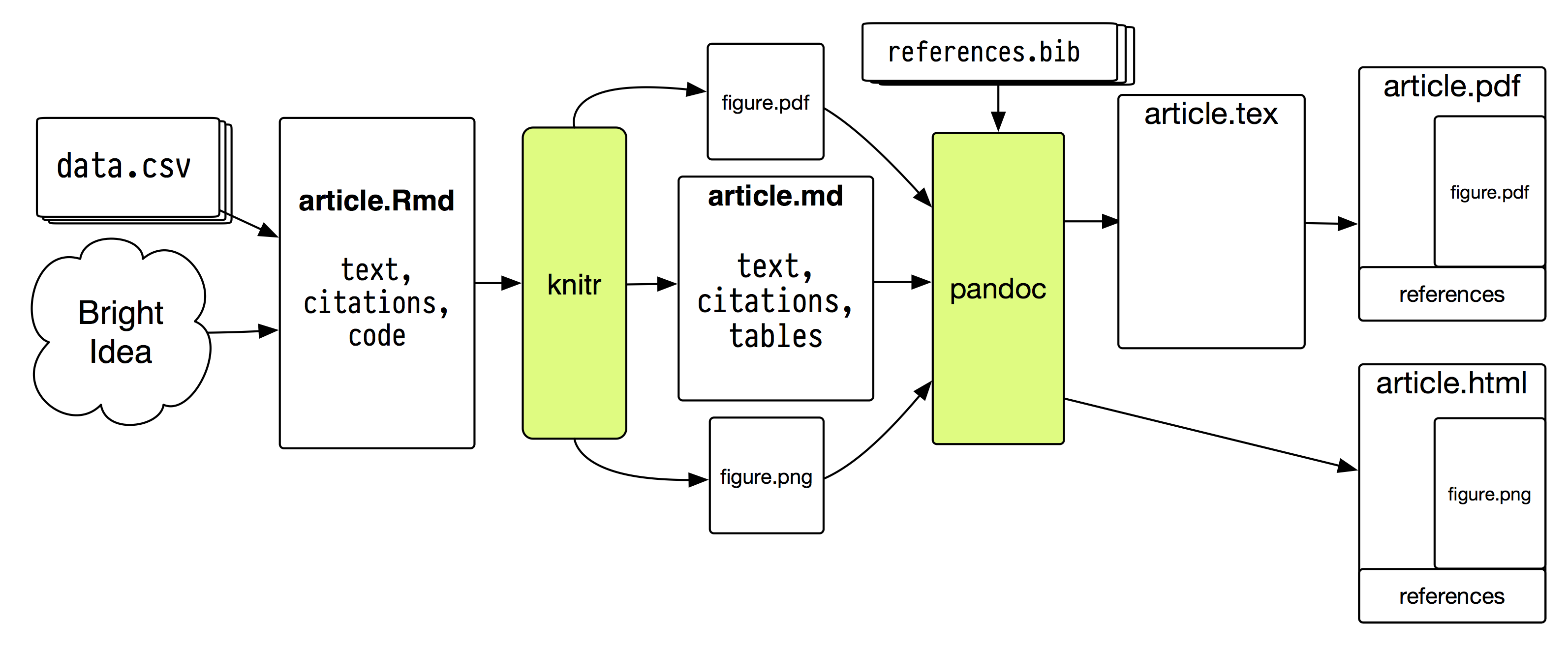
Figure 5.1: A plain-text document toolchain.
A sample document flow is shown in Figure 5.1. I promise it
is less insane than it appears. Describing it all at once might make
it sound a little crazy. But, at bottom, there are just two separable
pieces. First, knitr converts .Rmd files to .md files. Second,
John MacFarlane’s superb Pandoc converts .md
files to HTML, .tex, PDF, or Word formats. In both cases we use a
few switches, templates and configuration files to do that nicely and
with a minimum of fuss. You should install a standard set of Unix
developer tools, which on OS X can conveniently be installed
directly from the command line.8
along with R, knitr, pandoc, and a
TeX distribution. Note that the default
set-ups for knitr and pandoc—the two key pieces of the
process—will do most of what we want with no further tweaking. What
I will you here are just the relevant options to use and switches to
set for these tools, together with some templates and document samples
showing how nice-looking output can be produced in practice.
I write everything in Emacs, but as I hope is clear by now, that
doesn’t matter. Use whatever text editor you like and just learn the
hell out of it. The
custom LaTeX style files
were originally put together to let me write nice-looking .tex files
directly, but now they just do their work in the background. Pandoc
will use them when it converts things to PDF. The heavy lifting is
done by the
org-preamble-pdflatex.sty
and
memoir-article-styles
files. If you install these files where LaTeX can find them—i.e., if
you can compile a LaTeX document
based on this example—then
you are good to go. My
BibTeX master file is also
available, but you will probably want to use your own, changing
references to it in the templates as appropriate. Second, we have the
custom pandoc stuff.
Here is the repository for that.
Much of the material there is designed to go in the ~/.pandoc/
directory, which is where pandoc expects to find its configuration
files. I have also set up a sample
md-starter project and an
rmd-starter project. These
are the skeletons of project folders for a paper written in Markdown
(just an .md file, with no R) and a paper beginning life as an
.Rmd file. The sample projects contain the basic starter file and a
Makefile to produce .html, .tex, .pdf and .docx files.
Listing 2: Markdown file with document metadata
---
title: "A Pandoc Markdown Article Starter"
author:
- name: Kieran Healy
affiliation: Duke University
email: kjhealy@soc.duke.edu
- name: Joe Bloggs
affiliation: University of North Carolina, Chapel Hill
email: joebloggs@unc.edu
date: January 2014
abstract: "Lorem ipsum dolor sit amet."
...
# Introduction
Lorem ipsum dolor sit amet, consectetur adipisicing elit,
sed do eiusmod tempor incididunt ut labore et dolore magna
aliqua [@fourcade13classsituat]. Notice that citation.
# Theory
Lorem ipsum dolor sit amet, consectetur adipisicing
elit, sed do eiusmod tempor incididunt ut labore et
dolore magna aliqua [@fourcade13classsituat].Let’s start with a straightforward markdown file—no R code yet, so
nothing to the left of article.md line in Figure
5.1. The start of the sample
article-markdown.md file is shown in Listing 2. The bit at the
top is metadata, which pandoc understands. The HTML and LaTeX
templates
in the pandoc-templates repository are
set up to use this metadata properly.
For scholarly work, efficient management of citations is particularly
important. The pandoc-citeproc filter is an add-on that handles
these. It can be installed alongside pandoc. Your bibliography can
be stored in one of a variety of formats (such as BibTeX, or EndNote).
Within your .md document, cites are referred to by their key, such
as [@healy14datavisualsociol]. When pandoc converts your document,
the cite key is replaced with the reference information, like this
(Healy & Moody, 2014), and the full bibliographic entry is
included in an automaticaly-generated list of references.
Read
Pandoc’s documentation for more details about
citations. Pandoc can also handle in-text labels and cross-references.
A filter
named
pandoc-crossref
extends the extends the @label convention to deal to things like
figures, tables, equations, theorems, code listings and so on.
There is more than one way to have pandoc manage
citations, but here we just use the most self-contained route. Simple
documents can be contained in a single .md file. Documents including
data analysis start life as .Rmd files which are then knitted into
.md files and converted to PDF or HTML. At its simplest, a
mypaper.md document can be converted to mypaper.pdf file by
opening a terminal window and typing a command like the one in
Listing 3.
Listing 3: The simplest way to convert a Markdown file to PDF with pandoc
pandoc mypaper.md -o mypaper.pdf5.1 Automation with make
Because we will probably run commands like this a lot, it’s convenient to automate them a little bit, and to add some extra bells and whistles to accommodate things we will routinely add to files, such as author information and other metadata, together with the ability to process bibliographic information and cross-references. These are handled by pandoc by turning on various switches in the command, and by ensuring a couple of external “filters” are present to manage the bibliographies and cross-references. Rather than type long commands out repeatedly, we will automate the process. This kind of automation is especially useful when our final output file might have a number of prerequisites before it can be properly produced, and we would like the computer to be a little bit smart about what needs to be re-processed and under what conditions. That way, for example, if a Figure has not changed we will not re-run the (possibly time-consuming) R script to create it again, unless we have to.
We manage this process using a tool called
make. Inside our project
folder we put a plain-text Makefile that contains some rules
governing how to produce a target file that might have a number of
prerequisites. In this case, a PDF or HTML file is the target, and
the various figures and data tables are the prerequisites—if the
code that produces the prerequisites changes, the final document will
change too. Make starts from the final document and works backwards
along the chain of prerequisites, re-compiling or re-creating them as
needed. It’s a powerful tool. For a good basic introduction, take a
look at Karl Broman’s
“Minimal Make”. (Incidentally,
Karl Broman has
a number of tutorials and guides
providing accurate and concise tours of many of the tools and topics
described here, including
getting started with reproducible research,
using git and GitHub, and
working with knitr.)
Following Karl Broman’s example, let’s imagine that you have a paper,
paper.md written in Markdown, that incorporates references to a
figure, fig1.pdf generated by an R script, fig1.r. You could of
course have an .Rmd file that produces the output, but there are
situations where that isn’t ideal. The end-point or target is the full
article in PDF form. When the text of the paper changes in paper.md,
then paper.pdf will have to be re-created. In the same way, when we
change the content of fig1.r then fig1.pdf will need to be
updated, and thus also paper.pdf will need to be re-created as well.
Using make we can take care of the process.
Here is what a basic Makefile for our example would look like:
Listing 4: A simple Makefile
## Read as "mypaper.pdf depends on mypaper.md and fig1.pdf"
mypaper.pdf: mypaper.md fig1.pdf
pandoc mypaper.md -o mypaper.pdf
## Read as "fig1.pdf depends on fig1.r"
fig1.pdf: fig1.r
R CMD BATCH fig1.rThe big gotcha for Makefiles is that for no good reason they use the
tab key rather than spaces to indent the commands associated with
rules. If you use spaces, make will not work. With the Makefile in
Listing 4, typing make at the command line will have make
check the state of the final target (makefile.pdf) and all its
dependencies. If the target isn’t present, make will create it in
accordance with the rules specified. If the target is present,
make will check to see if any of its prerequisites have changed
since it was last created. If so, it will recreate the file. The chain
of prerequisites propagates backwards, so that if you change fig1.r,
then make will notice and re-run it to create fig1.pdf before
running the commands in the rule to create mypaper.pdf. You can also
choose to just invoke single rules from the makefile, e.g. by typing
make fig1.pdf instead of make at the command line. This will
evaluate just the fig1.pdf rule and any associated prerequisites.
For a simple example like this, make is mostly a minor convenience,
saving you the trouble of typing a sequence of commands over and over
to create your paper. However, it becomes very useful once you have
projects with many documents and dependencies—for example, a
dissertation consisting of separate chapters, each of which contains
figures and tables, which in turn depend on various R scripts to set
up and clean data. In those cases, make becomes a very powerful and
helpful way of ensuring your final output really is up to date.
To deal with more complex projects and chains of prerequisites, make
can make use of a number of variables to save you from (for example)
typing out the name of every figure-x.pdf in your directory.
The Makefile in the sample
md-starter project will
convert any markdown files in the working directory to HTML, .tex,
.pdf, or .docx files as requested. Typing make html at the
command line will produce .html files from any .md files in the
directory, for example. The PDF output (from make pdf) will look
like this article, more or less. The different pieces of the
Makefile define a few variables that specify the relationship
between the different file types. In essence the rules say, for
example, that all the PDF files in the directory depend on changes to
an .md file with the same name; that the same is true of the HTML
files in the directory, and so on. Then the show the pandoc commands
that generate the output files from the markdown input. The Makefile
itself is shown in Listing Listing 5 makes use of a few variables as
shorthand, as well as special variables like $@ and $<, which mean
“the name of the current target” and “the name of the current
prerequisite”, respectively.
Listing 5: A more complicated Makefile
## What extension (e.g. md, markdown, mdown) is being used
## for markdown files MEXT = md
## Expands to a list of all markdown files in the working directory
SRC = $(wildcard *.$(MEXT))
## Location of Pandoc support files.
PREFIX = /Users/kjhealy/.pandoc
## Location of your working bibliography file
BIB = /Users/kjhealy/Documents/bibs/socbib-pandoc.bib
## CSL stylesheet (located in the csl folder of the PREFIX directory).
CSL = apsa
## x.pdf depends on x.md, x.html depends on x.md, etc
PDFS=$(SRC:.md=.pdf)
HTML=$(SRC:.md=.html)
TEX=$(SRC:.md=.tex)
DOCX=$(SRC:.md=.docx)
## Rules -- make all, make pdf, make html. The `clean` rule is below.
all: $(PDFS) $(HTML) $(TEX) $(DOCX)
pdf: clean $(PDFS)
html: clean $(HTML)
tex: clean $(TEX)
docx: clean $(DOCX)
## The commands associated with each rule. The first one is run with
## make html.
%.html: %.md
pandoc -r markdown+simple_tables+table_captions+yaml_metadata_block \
-w html -S --template=$(PREFIX)/templates/html.template \
--css=$(PREFIX)/marked/kultiad-serif.css --filter pandoc-crossref \
--filter pandoc-citeproc --csl=$(PREFIX)/csl/$(CSL).csl \
--bibliography=$(BIB) -o $@ $<
## Same goes for the other file types.
## Watch out for the TAB before 'pandoc'
%.tex: %.md
pandoc -r markdown+simple_tables+table_captions+yaml_metadata_block \
--listings -w latex -s -S --latex-engine=pdflatex \
--template=$(PREFIX)/templates/latex.template \
--filter pandoc-crossref --filter pandoc-citeproc \
--csl=$(PREFIX)/csl/ajps.csl --filter pandoc-citeproc-preamble \
--bibliography=$(BIB) -o $@ $<
%.pdf: %.md
pandoc -r markdown+simple_tables+table_captions+yaml_metadata_block \
--listings -s -S --latex-engine=pdflatex \
--template=$(PREFIX)/templates/latex.template \
--filter pandoc-crossref --filter pandoc-citeproc \
--csl=$(PREFIX)/csl/$(CSL).csl --filter pandoc-citeproc-preamble \
--bibliography=$(BIB) -o $@ $<
%.docx: %.md
pandoc -r markdown+simple_tables+table_captions+yaml_metadata_block \
-s -S --filter pandoc-crossref --csl=$(PREFIX)/csl/$(CSL).csl \
--bibliography=$(BIB) -o $@ $<
clean:
rm -f *.html *.pdf *.tex *.aux *.log *.docx
.PHONY: cleanNote that the pandoc commands are interpreted single lines of text,
not several lines separated by the return key. But you can use the
\ symbol to tell make to continue to the next line without a
break. With this Makefile, typing make pdf would take all the .md
files in the directory one at a time and run the pandoc command to
turn each one into a PDF, using the
APSR
reference style, my latex template, and a .bib file called
socbib-pandoc.bib.
You shouldn’t use this Makefile blindly. Take the time to learn how
make works and how it can help your project. The
official manual is pretty
clear. Make’s backward-looking chain of prerequisites can make it
tricky to write rules for complex projects. When writing or inspecting
a Makefile and its specific rules, it can be helpful to use the
--dry-run switch, as in make --dry-run. This will print out the
sequence of commands make would run, but without actually executing
them. You can try this with the Makefile in Listing 5 in a
directory with at least one .md file in it. For example, look at the
commands produced by make pdf --dry-run or make docx --dry-run or
make clean --dry-run.
The particular steps needed for many projects may be quite simple, and
not require the use of any variables or other frills. If you find
yourself repeatedly running the same sequence of commands to assemble
a document (e.g. cleaning data; running preliminary code; producing
figures; assembling a final document) then make can do a lot to
automate the process. For further examples of Makefiles doing things relevant to data analysis, see Lincoln Mullen’s discussion of the things he uses make to manage.
The examples directory
includes
a sample .Rmd file. The code chunks in the file provide examples of
how to generate tables and figures in the document. In particular they
show some useful options that can be passed to knitr.
Consult the knitr project page for
extensive documentation and many more examples. To produce output from
the article-knitr.Rmd file, you could of course launch R in the
working directory, load knitr, and process the file. This produces
the article-knitr.md file, together with some graphics in the
figures/ folder (and some working files in the cache/ folder). We
set things up in the .Rmd file so that knitr produces both PNG and
PDF versions of whatever figures are generated by R. That prepares the
way for easy conversion to HTML and LaTeX. Once the article-knitr.md
file is produced, HTML, .tex, and PDF versions of it can be produced
as before, by typing make at the command line. But of course there’s
no reason make can’t automate that first step, too. The
rmd-starter project has a
sample Makefile that begins with the .Rmd files in the directory
and produces the outputs from there.